Custom Eprobe®/Eprimer™ Synthesis : Technology Overview
□Technical Information
Eprobe®, which was jointly developed by DNAFORM and RIKEN, is a synthetic oligonucleotide-based fluorescent probe that has an excitonic interaction. Performing polymerase chain reaction (PCR) with Eprobe® allows for specific quantification of target DNA sequences and also enables base mutation detection.
□Features of Eprobe®
- □Eprobe® generates a fluorescence signal upon hybridization to its target nucleotide sequence.
- □The use of a single Eprobe® allows for both real-time PCR and melting curve analysis.
- □Higher melting temperature (Tm values) compared to normal DNA oligonucleotides are observed.
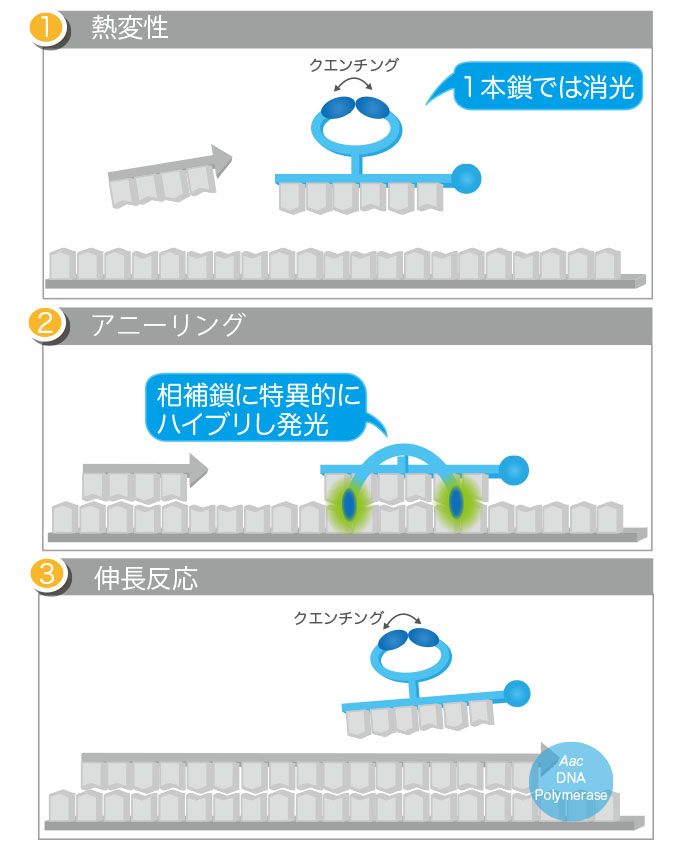
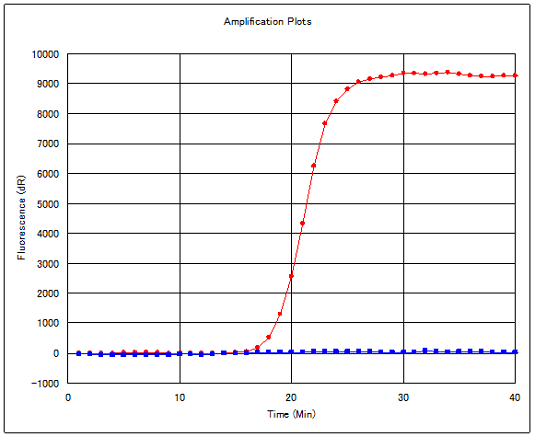
□Function of Eprobe® in real-time PCR
Eprobe® is a fluorescence-labeled oligonucleotide with one modified thymine carrying two dye moieties. When Eprobe® forms a double-stranded structure with target DNA sequences, the two dyes in Eprobe® are separated and emit a strong fluorescence caused by disruption of the excitonic interaction between dyes. Eprobe®, when added in sufficient amounts to the reaction mixture for real-time PCR, also binds to its target DNA sequence during the primer annealing step. By measuring the fluorescence generated upon binding of Eprobe®, only the target DNA sequence is quantified.
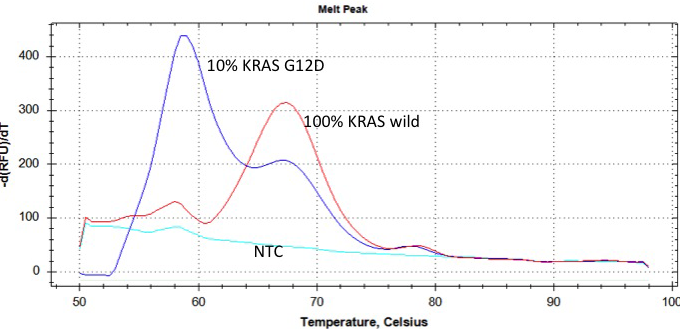
This Eprobe®-mediated real-time PCR can also be applied to the detection of base mutations. In post-PCR melting curve analysis, differences in melting temperature (Tm) between when Eprobe® binds to a perfectly matched wild-type base sequence and when the same Eprobe® binds to mismatched mutated base sequence(s) are expected. By analyzing the differences in Tm, the presence or absence of mutated DNA sequence(s) can be assessed.
□Usage of Eprobe®
When Eprobe® binds to a perfectly matched wild-type base sequence and when the same Eprobe® binds to mismatched mutated base sequence(s), differences in Tm are observed in post-PCR melting curve analysis. If differences in Tm are analyzed, the presence or absence of variations in DNA sequences can be determined.
□Eprobe® & Eprimer™
□Eprobe®

Eprobe® is modified at the 3’-end with a C3 spacer to prevent primer extension during PCR, which ensures high reaction specificity when using Eprobe® as the PCR probe.
□Eprimer™

Eprimer™ has no modifications at the 3’-end, which allows Eprimer™ to undergo normal primer extension from the 3’-end during PCR.
□Typical conditions
Concentrations of primers and Eprobe® for use in Eprobe®-mediated PCR
The PCR primer for annealing to the strand complementary to Eprobe® (the strand to which Eprobe® will bind) is used at a concentration of 0.1 μM, while the PCR primer for annealing to the strand to which Eprobe® will not bind is used at 0.5 μM. The concentration of Eprobe® is set at 0.4 μM.
□Typical Eprobe® PCR condition
| Temperature | Time | Cycles |
|---|---|---|
| 95℃ | 10 min | |
| 95℃ | 15 sec | 50 cycles |
| 58℃ | 30 sec | |
| 72℃ | 12 sec |
□Typical condition for melting curve analysis with Eprobe® PCR
| Temperature | Time |
|---|---|
| 37℃ | 7 min |
| 95℃ |
□Calculation of Tm in Eprobe®-mediated PCR
When introduced into Eprobe® (or Eprimer™), thiazole orange serves to increase the Tm of the hybridization product ? i.e., Eprobe®/DNA (or Eprimer™/DNA) duplex. The predicted Tm for Eprobe® (or Eprimer™) can be calculated using an online tool (available on the following website) by placing “Z” in the position of thiazole orange-labeled thymidine in the oligonucleotide.
→RIKEN > Hybridization thermodynamics of ECHO/DNA duplexes
*The data available on the website is based on publications by RIKEN.